Kegg Chart Examples
Kegg Chart Examples - Web citations (294) references (14) figures (1) abstract and figures. This example is on vitis vinifera (as the prefix vvi can suggest), but the approach is general. In this vignette, we demonstrate the application of kegggraph as exible module in analysis pipelines targeting heterogenous biological questions. Web kegggraph is an interface between kegg pathway and graph object as well as a collection of tools to analyze, dissect and visualize these graphs. It parses the regularly updated kgml (kegg xml) files into graph models maintaining all. Global/overview carbohydrate energy lipid nucleotide amino acid other amino glycan. Web kegg pathway is a collection of manually drawn pathway maps representing our knowledge of the molecular interaction, reaction and relation networks for: It parses the regularly updated kgml (kegg xml) files into graph models maintaining all essential pathway attributes. Kegggraph is an interface between kegg pathway and graph object as well as a collection of tools to analyze, dissect and visualize these graphs. But, have you ever wondered what is happening deep down in the kegg brains, eh, i mean kegg sensing technology and predictive algorithms? Kegg pathway is a service of kyoto encyclopedia of genes and genomes (kegg), constructing manually curated pathway maps that. Conversion of kegg ids, ec numbers and cog ids to kegg orthologs (ko) and of ko to ec numbers. Keggpathwaygraphs(organism = hsa, targreltypes = c(gerel, pcrel, pprel), relpercthresh = 0.9, nodeonlygraphs = false, updatecache = false, verbose =. It parses the. Kegggraph is an interface between kegg pathway and graph object as well as a collection of tools to analyze, dissect and visualize these graphs. The chart consists of various nodes and edges, each representing different biological entities and their interactions. Web description kegggraph is an interface between kegg pathway and graph object as well as a collection of tools to. The first step in each topgo analysis is to create a topgodata object. Web kegg pathway is a collection of manually drawn pathway maps representing our knowledge of the molecular interaction, reaction and relation networks for: Download and parse kegg pathway data. It parses the regularly updated kgml (kegg xml) files into graph models maintaining all. Web in this regard,. Web citations (294) references (14) figures (1) abstract and figures. Conversion of kegg ids, ec numbers and cog ids to kegg orthologs (ko) and of ko to ec numbers. Cofactor/vitamin terpenoid/pk other secondary metabolite xenobiotics. Kegggraph also downloads kegg pathway information and converts it into a format that can be effectively analyzed in r (zhang and. This example is on. In this article we’ll share what to look for. Kegggraph is an interface between kegg pathway and graph object as well as a collection of tools to analyze, dissect and visualize these graphs. The ko assignment data are linked to kegg mapper (kanehisa et al., 2022) for more detailed analysis. Web kegggraph is an interface between kegg pathway and graph. We demonstrate the capabilities of the kegggraph package, an interface between kegg pathways and graph model in r as well as a collection of tools for these graphs. Keggsum receives a kegg identifier (kid) as an input, connects to the kegg database, downloads a specialized form of the pathway, and determines the most important nodes in the graph. Let’s consider. In this vignette, we demonstrate the application of kegggraph as exible module in analysis pipelines targeting heterogenous biological questions. It parses the regularly updated kgml (kegg xml) files into graph models maintaining all. We demonstrate the capabilities of the kegggraph package, an interface between kegg pathways and graph model in r as well as a collection of tools for these. Begin by identifying the nodes in the chart. In this article we’ll share what to look for. Web citations (294) references (14) figures (1) abstract and figures. The other pie charts show taxonomic distributions of proteins in the sample for cellular organisms (figure 4b) and viruses. It parses the regularly updated kgml (kegg xml) files into graph models maintaining all. We demonstrate the capabilities of the kegggraph package, an interface between kegg pathways and graph model in r as well as a collection of tools for these graphs. Web citations (294) references (14) figures (1) abstract and figures. Web as a useful note to self, i paste here an easy example on the use of the pathview package by weijun. Web citations (294) references (14) figures (1) abstract and figures. Conversion of kegg ids, ec numbers and cog ids to kegg orthologs (ko) and of ko to ec numbers. Web as a useful note to self, i paste here an easy example on the use of the pathview package by weijun luo to plot the log fold change of gene. The other pie charts show taxonomic distributions of proteins in the sample for cellular organisms (figure 4b) and viruses. This example is on vitis vinifera (as the prefix vvi can suggest), but the approach is general. Let’s consider a sample kegg chart that represents a biological pathway related to a specific disease. We demonstrate the capabilities of the kegggraph package, an interface between kegg pathways and graph model in r as well as a collection of tools for these graphs. Kegggraph also downloads kegg pathway information and converts it into a format that can be effectively analyzed in r (zhang and. We demonstrate the capabilities of the kegggraph package, an interface between kegg pathways and graph model in r as well as a collection of tools for these graphs. In this vignette, we demonstrate the application of kegggraph as exible module in analysis pipelines targeting heterogenous biological questions. Kegggraph is an interface between kegg pathway and graph object as well as a collection of tools to analyze, dissect and visualize these graphs. Cofactor/vitamin terpenoid/pk other secondary metabolite xenobiotics. Kegg pathway is a service of kyoto encyclopedia of genes and genomes (kegg), constructing manually curated pathway maps that. Web kegg pathway is a collection of manually drawn pathway maps representing our knowledge of the molecular interaction, reaction and relation networks for: Web as a useful note to self, i paste here an easy example on the use of the pathview package by weijun luo to plot the log fold change of gene expression across a given kegg pathway. Begin by identifying the nodes in the chart. The chart consists of various nodes and edges, each representing different biological entities and their interactions. The first step in each topgo analysis is to create a topgodata object. Web the first pie chart (figure 4a) is a summary of ko assignment shown in kegg functional categories.The marked KEGG metabolic pathway map by red points with all the

The bar charts for gene ontology analysis and KEGG pathway analysis for

KEGG pathway analysis. The bubble chart shows the KEGG pathway analysis
KEGG pathway analysis. (a) KEGG pathway enrichment analysis was

How to generate KEGG pathway classification chart

GO term and KEGG pathway enrichment analysis of DEGs. (a,b) GO terms
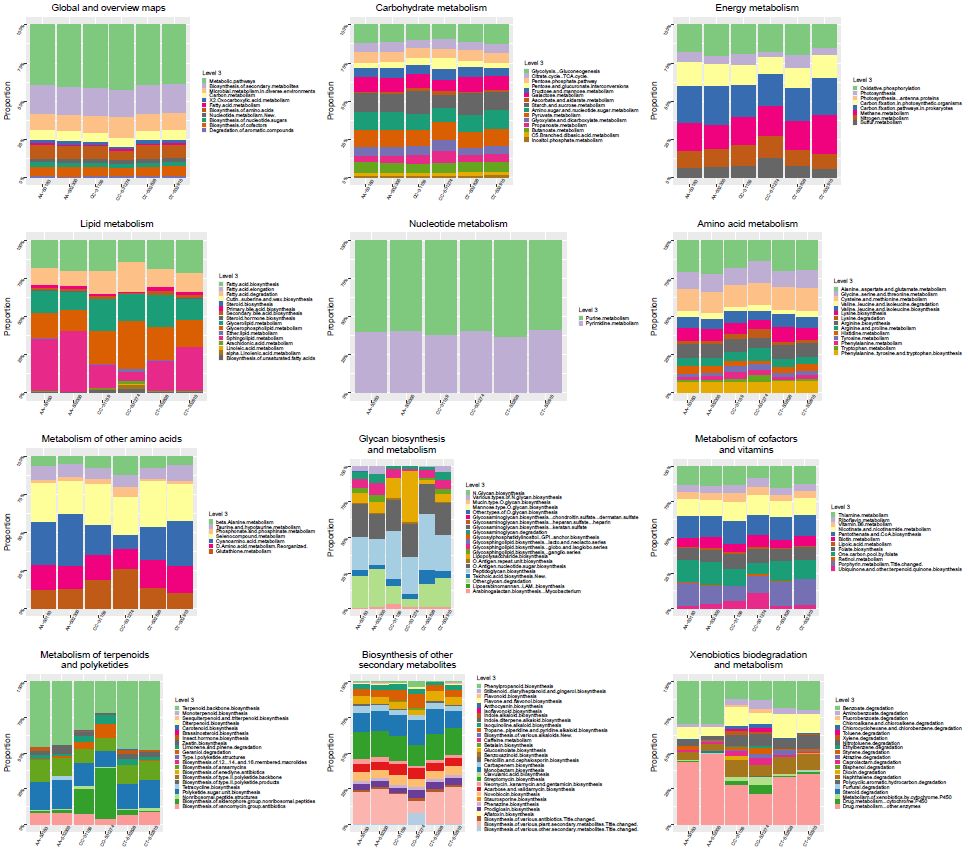
KEGG解析の結果を可視化「KEGG pathway classification chart」 株式会社生物技研

Kegg Pathway Enrichment Analysis KEGG pathway enrichment analysis for

KEGG pathway analysis of identified DEGs from contralateral DRG. (A

KEGG pathway analysis of differentially expressed genes. Advanced
Conversion Of Kegg Ids, Ec Numbers And Cog Ids To Kegg Orthologs (Ko) And Of Ko To Ec Numbers.
For Basic Use Of The Kegggraph Package, Please Refer To The Vignette Kegggraph:
Web Description Kegggraph Is An Interface Between Kegg Pathway And Graph Object As Well As A Collection Of Tools To Analyze, Dissect And Visualize These Graphs.
Download And Parse Kegg Pathway Data.
Related Post: